| Gene Symbol | Tsp_12822 |
| Entrez Gene ID | 10906439 |
| Full Name | DNA mismatch repair protein Msh6 |
| Gene Type | protein-coding |
| Organism | Trichinella spiralis |
HOME
ORF » Species Summary » Trichinella spiralis » Tsp_12822 cDNA ORF clone
| Gene Symbol | Tsp_12822 |
| Entrez Gene ID | 10906439 |
| Full Name | DNA mismatch repair protein Msh6 |
| Gene Type | protein-coding |
| Organism | Trichinella spiralis |
| mRNA | Protein | Name |
|---|---|---|
| XM_003367841.1 | XP_003367889.1 | DNA mismatch repair protein Msh6 |


The draft genome of the parasitic nematode Trichinella spiralis.
Mitreva M, Jasmer DP, Zarlenga DS, Wang Z, Abubucker S, Martin J, Taylor CM, Yin Y, Fulton L, Minx P, Yang SP, Warren WC, Fulton RS, Bhonagiri V, Zhang X, Hallsworth-Pepin K, Clifton SW, McCarter JP, Appleton J, Mardis ER, Wilson RK
Nature genetics43(3)228-35(2011 Mar)
GeneRIFs: Gene References Into Functions What's a GeneRIF?
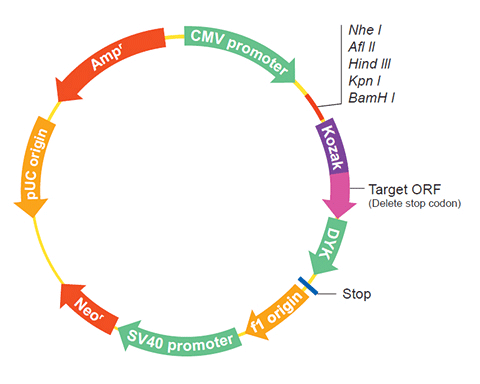
The following Tsp_12822 gene cDNA ORF clone sequences were retrieved from the NCBI Reference Sequence Database (RefSeq). These sequences represent the protein coding region of the Tsp_12822 cDNA ORF which is encoded by the open reading frame (ORF) sequence. ORF sequences can be delivered in our standard vector, pcDNA3.1+/C-(K)DYK or the vector of your choice as an expression/transfection-ready ORF clone. Not the clone you want? Click here to find your clone.
| CloneID | OTr121743 | |
| Clone ID Related Accession (Same CDS sequence) | XM_003367841.1 | |
| Accession Version | XM_003367841.1 Latest version! | Documents for ORF clone product in default vector |
| Sequence Information | ORF Nucleotide Sequence (Length: 1059bp) Protein sequence SNP |
|
| Vector | pcDNA3.1-C-(k)DYK or customized vector |  User Manual User Manual |
| Clone information | Clone Map |  MSDS MSDS |
| Tag on pcDNA3.1+/C-(K)DYK | C terminal DYKDDDDK tags | |
| ORF Insert Method | CloneEZ™ Seamless cloning technology | |
| Insert Structure | linear | |
| Update Date | 2011-06-30 | |
| Organism | Trichinella spiralis | |
| Product | DNA mismatch repair protein Msh6 | |
| Comment | Comment: PROVISIONAL REFSEQ: This record has not yet been subject to final NCBI review. This record is derived from an annotated genomic sequence (NW_003530522). Trichinella spiralis is a roundworm that cause most of the human trichinella infections and deaths around the world. Its pathogenicity is higher than that of other trichinella species due to the higher number of newborn larvae produced by the females and for the stronger immune reaction induced in humans. The life cycle of the parasite begins when a person or an animal eats contaminated meat containing larvae. T. spiralis is a basal nematode with a well-defined phylogenetic position near the root of the phylum Nematoda. The genome size estimate based on flow sorted nuclei stained with PI (Spencer Johnston, Texas A&M University) is 1C = 71.3 +/1 1.2 Mb. The strain being sequenced (ISS 195) was obtained from the laboratory of Judith Appleton (Cornell University) and has been maintained in rats since 1970. Worm isolation and DNA extraction was performed by Dante Zarlenga (USDA) This assembly consists of plasmid, fosmid and BAC end sequences. The data were assembled using the assembly engine, PCAP (Xiaoqiu Huang et. al. 2006). Our goal is to explore this WGS draft sequence of T. spiralis in several ways: i) to provide a better understanding of evolutionary biology by identifying gene loss or gain across the phylum Nematoda and clarify evolution of genome architecture (synteny, operons); ii) help identify RNA genes and regulatory regions; and iii) better define proteins involved in nematode parasitism that impact health and disease and are relevant to both host-parasite relationships and basic biological processes.. We masked the repeats by using RECON (Bao and Eddy, 2002) and RepeatMasker (A.F.A. Smit, R. Hubley & P. Green RepeatMasker at http://repeatmasker.org). Then the Ribosomal RNA genes were identified using RNAmmer ((http://www.cbs.dtu.dk/cgi-bin/nph-sw_request?rnammer ). Transfer RNA genes were identified with tRNAscan-SE (Lowe and Eddy, 1997). Non-coding RNAs, such as microRNAs, were identified by sequence homology search of the Rfam database (http://selab.janelia.org/software.html). Protein-coding genes were predicted using a combination of ab initio programs (Snap, Korf, 2004 and Fgenesh, Softberry, Corp) and an inhouse evidence based program Eannot (Eannot Ding et al., 2004) which uses mRNA, EST and protein alignment information from same species or cross-species to aid in gene structure determination. A consensus gene set from the above prediction algorithms will be generated, using a logical, hierarchical approach. Gene product naming was determined by BER (JCVI: http://ber.sourceforge.net ). For information regarding this assembly or project, or any other GSC genome project, please visit our Genome Groups web page (http://genome.wustl.edu/genome_group_index.cgi) and email the designated contact person. For specific questions regarding the T. spiralis genome project contact Makedonka Mitreva [email protected] (Washington University School of Medicine). The National Human Genome Research Institute (NHGRI) of the National Institutes of Health (NIH) provided funds for this project. | |
1 | ATGAGTTCGA TTTTCGCTCG CAAAGGGAAG ATCGATAGGC TTCAAAAGGT ACACGACAAA |
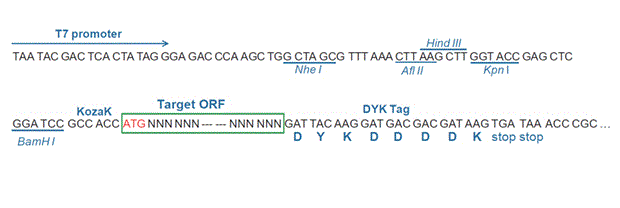
The stop codons will be deleted if pcDNA3.1+/C-(K)DYK vector is selected.
| RefSeq | XP_003367889.1 |
| CDS | 1..1059 |
| Translation |

Target ORF information:
Target ORF information:
|
 XM_003367841.1 XM_003367841.1 |
1 | ATGAGTTCGA TTTTCGCTCG CAAAGGGAAG ATCGATAGGC TTCAAAAGGT ACACGACAAA |
The stop codons will be deleted if pcDNA3.1+/C-(K)DYK vector is selected.
The draft genome of the parasitic nematode Trichinella spiralis. |